Resources
Conducting reproducible research
Conducting reproducible research is important for large projects and error checking. Reproducible research allows people reproducing your published results easily and it is commonly required for publishing. Large-scale reproducible research includes an organized pipeline suitable for high-throughput computing and clear documentation. Keeping your pipeline and folders organized improves efficiency and helps avoiding potential confusion in the future. Documenting your work is like leaving messages to yourself in the future. Having a clear and up-to-date documentation allows you to keep track of your work and eliminate the hustle and bustle. Finally, making your pipeline suitable for high throughput computing greatly improves the speed of the analysis but you should be very careful about the errors. Handling a huge batch of tasks may cause error, performing defensive programming and designing an efficient error detection protocol help you eliminate the errors.
Here, I am going to provide some general advice on organizing the pipeline, documenting the work, and scaling up the pipeline. The advice is based on my research experience at UW-Madison and focuses on genome-wide association studies (GWAS). Those are the things I wish I knew when I joined the lab, and I hope the advice is helpful for you.
1. Organizing your pipeline
Organize your pipeline by the type and usage of your data. Typically, I start my project directory like this.
Project
├── Data # for inputs
├── Results # for outputs and results)
├── Plots # for plot outputs
└── Scripts # for storing scripts
├── 1-1.xxx.sh # the script for step 1-1
├── 1-2.xxx.sh # the script for step 1-2
└── ...
Run your scripts under Scripts and store the output files to Results and Plots. Start the script names by the number of the corresponding step so that the scripts will be ordered accordingly. Remember to use relative path in your scripts so that the codes are still executable after changing the path of project directory.
Follow a consistent folder organization and naming tradition through your project. This saves you a lot of time when you revisit your pipeline.
2. Documenting your work
I include at least these information in my documentation.
- Major changes of the pipeline and the dates
- The summary of the pipeline
- The input/output file formats and their path
- A detailed explanation of each step
Here is the template I used to document my work.
3. Working on high throughput clusters
Software
TITANS
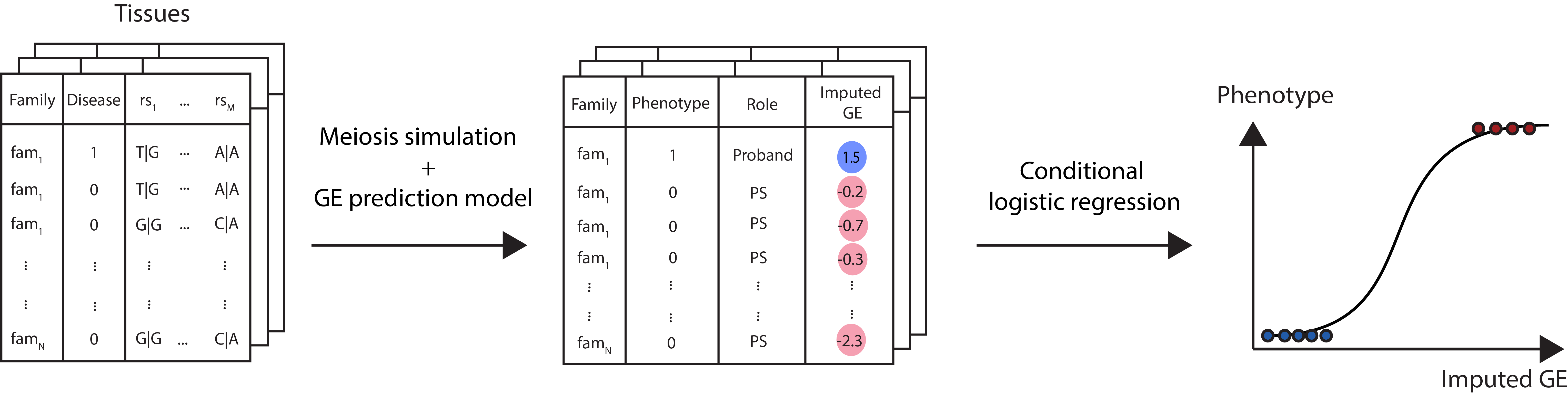
TITANS (TrIo-based Transcriptome-wide AssociatioN Study) is a statistical framework for conducting transcription-wide associaiton study in proband-parent trios. TITANS quantifies the transmission disequilibrium of genetically regulated gene expression from parents to probands using pseudosibling matching and conditional logistic regression.
